珊瑚礁作为热带、亚热带海洋鱼类重要的产卵、育幼场,受到广泛关注。但是由于人类活动以及全球气候变化影响,世界范围内的珊瑚礁生态系统受到了严重威胁[1]。例如世界最大珊瑚礁群——澳大利亚大堡礁的活珊瑚覆盖率已由1960年的50%下降到2003年的20%[2];加勒比海的活珊瑚覆盖率由1977年的50%下降到2001年的10%[3]。中国南海珊瑚礁中,广东大亚湾的活珊瑚覆盖率由1983—1984年的76.6%下降到2008年的15.3%[4];海南三亚鹿回头的活珊瑚覆盖率由1960年的80%~90%下降到2009年的12%[5];西沙永兴岛由1980年的90%下降到2008年的10%[6]。珊瑚礁生态系统的严重退化,势必威胁到依靠其栖息和繁育的海洋鱼类。珊瑚礁生态系统的科学管理与保护,日益受到世界范围内海洋环保工作者的重视。
徐闻珊瑚礁(20°10'N~20°27'N)位于广东省雷州半岛西南部,从徐闻西连一直分布到角尾南岭海域,是中国大陆珊瑚礁分布的最北端,也是大陆沿岸面积最大、种类最多、唯一发育和保存最完好的珊瑚岸礁[7]。徐闻国家级珊瑚礁保护区(面积约109 km2)即位于此。徐闻珊瑚礁海域鱼类资源十分丰富,曾报道鱼类135种[8]。该海域亦是鱼类重要的产卵、育幼场,但未见有关该海域的鱼卵、仔稚鱼相关研究的报道。近年来,随着全球气候变化和环境污染的加剧,徐闻珊瑚礁保护区活珊瑚覆盖率仅为2005年的1/3,栖息地生境退化十分严重,已影响到其对鱼类的产卵、育幼功能。因此,对徐闻珊瑚礁海域鱼类的产卵育幼场、产卵亲体和幼鱼资源的保护工作迫在眉睫。摸清该海域鱼卵、仔稚鱼的种类组成与分布是开展珊瑚礁鱼类资源保护与恢复工作的重要前提。
鱼类早期生活史阶段作为鱼类生活史中的一个重要环节,其成活率的高低直接关系到年际补充量的大小,是引起种群数量变动和年龄结构变化的主要原因[9]。由于仔稚鱼形态特征复杂多变,可用于鉴定的可识别的形态特征相对较少,因而基于传统形态分类学的鉴定往往在准确性方面存在较大的局限性[10]。南海海洋鱼类超过3 300种,但是可参考的仔稚鱼形态学图鉴种类不足300种,加之传统的形态学鉴定需要长期训练和很丰富的鉴定经验,因此寻求更为稳定、可靠的分类鉴定方法,是开展鱼类早期资源研究的关键[11-12]。DNA条形码技术,即利用线粒体细胞色素 C 氧化酶亚基 Ⅰ(Mitochondrial cytoehrome coxidase subunit Ⅰ,CO Ⅰ)基因的一段约 648 bp长度的基因片段进行物种鉴定的技术,因其鉴定快速、操作便捷和较为可信,为仔稚鱼的分类鉴定研究创造了机遇[13⇓⇓-16]。已有许多研究表明,DNA条形码是一种鉴定仔稚鱼的较为可靠的方法,例如Hubert N等[13]应用DNA条形码成功鉴定了莫雷阿岛的373尾仔稚鱼;Valdez-Moreno M等[14]结合成鱼DNA条形码鉴定了墨西哥尤卡坦半岛的仔稚鱼;Hou G等[17]基于DNA条形码技术鉴定了南海101种甲醛保存的仔稚鱼。此外,截至2023年5月,BOLD数据库中已经包含了24 776种鱼类439 913个序列,为仔稚鱼的分子鉴定提供了便利和参考[18]。
为更准确地了解徐闻珊瑚礁海域仔稚鱼种类组成,本研究选取了2018—2020年徐闻角尾海域夏、秋季的仔稚鱼为研究对象,根据形态学初步筛选部分仔稚鱼样本进行扩增和测序,通过与公共数据库的匹配来实现仔稚鱼的分子鉴定,并探讨了DNA条形码技术在徐闻珊瑚礁海域仔稚鱼分类中的适用性,以期为徐闻珊瑚礁海域的产卵育幼场保护和渔业资源的可持续利用提供数据支撑和理论依据。
1 材料与方法
1.1 数据来源
仔稚鱼样品来自徐闻角尾海域的2018—2020年度夏、秋季鱼类浮游生物调查。采样工具为定置网(网口长4.0 m、宽1.5 m,网目大小为1.0 mm)和大型浮游生物网(长2.70 m,网口直径为0.80 m,网目大小为0.505 mm)。采样时期选定调查月份的大潮时期,浮游生物网采集时间为05:00~17:00,采集白天的鱼卵、仔稚鱼(站位S1~S3);定置网的下网时间为17:00~翌日05:00,利用潮汐采集晚上的仔稚鱼(站位S4~S5)。使用浮游生物网进行水平拖网调查,拖时为10 min,拖速为1.5~2.2节。水平拖曳时,于浮游生物网上方悬挂适量的浮子,以保证拖曳时网口既不沉入水底又能刚好完全没入水中。样品采集后一部分迅速固定于75%乙醇溶液保存,另一部分固定在4%甲醛溶液保存,然后带回实验室。具体采样时间和站位如表1所示。
表1 徐闻角尾海域具体采样时间和站位
Tab.1
| 采样时间 Sampling time | 站位 Stations | 经纬度 Latitude and longitude | 水深/m Depth |
|---|---|---|---|
| 2018-10-27—10-28 | S4、S5 | N 20°11'~20°13' E 109°59'~110°00' | 10~15 |
| 2019-08-27—08-28 | S1-S5 | N 20°11'~20°15' E 109°56'~110°00' | 4~15 |
| 2019-09-14 | S1-S5 | 4~15 | |
| 2020-08-04 | S1-S5 | 4~15 | |
| 2020-09-28 | S1-S5 | 4~15 |
1.2 DNA提取与目的片段扩增
将乙醇溶液保存的仔稚鱼逐一挑选出后,首先按照形态进行初步鉴定,对准备DNA提取的样品进行编号,用蔡司体式显微镜(ZEISS SteREO Discovery.V20)拍照,然后每个样本独立放入装有预冷过的75%乙醇溶液的2 mL离心管中备用。使用预冷的超纯水中洗脱酒精约5 min,使用UE基因组DNA小量制备试剂盒(优逸兰迪,苏州)提取DNA,置于-20 ℃下保存。鱼类CO Ⅰ基因片段扩增通用引物:F1:5’-TCAACCAACCACAAAGACATTGGCAC-3’和FishR1:5’-TAGACTTCTGGGTGGCCAAAGAATCA-3’[19]。PCR扩增总体积为40 μL,包括20 μL Tsingke TM Master Mix、每种引物1 μL(10 pmol)、1~10 μL总DNA模板和8~17 μL ddH2O(总DNA模板+ ddH2O =18 μL)。PCR条件为:94 ℃预变性3 min;94 ℃ 变性30 s, 51 ℃退火30 s, 72 ℃延伸1 min,35个循环;最后72 ℃延伸10 min[17]。PCR产物经1.2%琼脂糖凝胶电泳检测合格后送武汉天一辉远双向测序。
1.3 数据分析
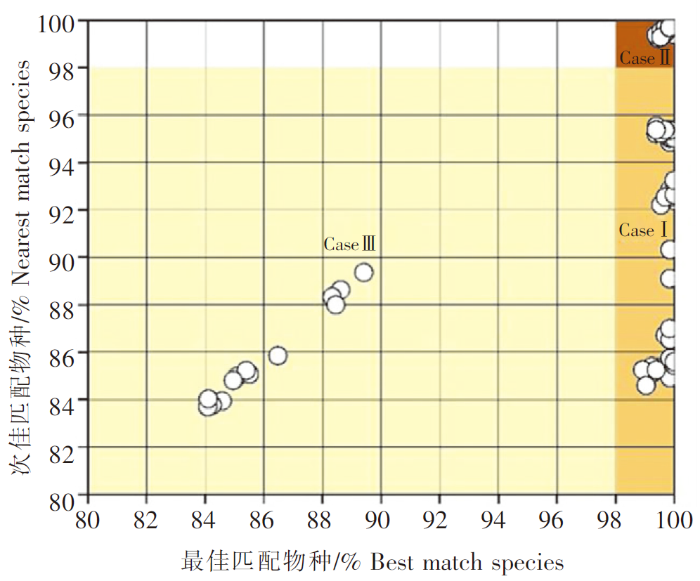
获取双向测通的测序结果后,通过Lasergene 7.0中的SeqMan对序列进行拼接,去除两端引物,获得长度为655 bp左右的COⅠ序列。将COⅠ序列与BOLD系统(BOLD,V4)中的序列进行匹配度层次分析,分析时仅选择已公布登入号(Genbank)的物种序列。在前100匹配度序列最高的物种中,分别界定为最佳匹配物种(Best match species)和次佳匹配物种(Nearest match species),记录对应鱼种的学名、匹配相似度以及序列号。在前100个匹配度序列中,无次佳匹配物种时记为null。根据序列与最佳匹配物种和次佳匹配物种的遗传相似度,当最佳匹配物种相似性>98%和次佳匹配物种相似性≤98%或null时,将仔稚鱼序列界定为Case Ⅰ;当最佳匹配物种相似性>98%和次佳匹配物种相似性>98%时,将仔稚鱼序列界定为Case Ⅱ;当最佳匹配相似性≤98%和次匹配物种相似性≤98%时,将仔稚鱼序列界定为Case Ⅲ[13]。使用Mega v7.0软件检查简并碱基,删除低质量的序列,基于K2P(Kimura-2-parameter)模型[20],采用邻接法(Neighbor joining method,NJ法)构建系统进化树来验证比对结果。
2 结果与分析
2.1 样品序列扩增与分析
本研究共测序了236尾徐闻角尾珊瑚礁海域的仔稚鱼,双向测通COⅠ序列204个,去除低质量的5个序列后,最终获得199个高质量的COⅠ序列,占比总测序的84.32%,纳入后续分析。使用98%相似性阈值界定,有89.95%序列(179/199)的最佳匹配度在98%~100%之间;次佳匹配的相似性在0%~100%之间。在199个仔稚鱼序列中,有69个序列,归属于Case Ⅰ,占总比为34.67%;有110个序列,归属于Case Ⅱ,占总比为55.28%;有20个仔稚鱼序列,归属于Case Ⅲ,占总比为10.05%(图1)。
图1
图1
仔稚鱼匹配分布散点图
Fig.1
The scatter plot of matching degree of fish larvae and juvenile
2.2 种类鉴定
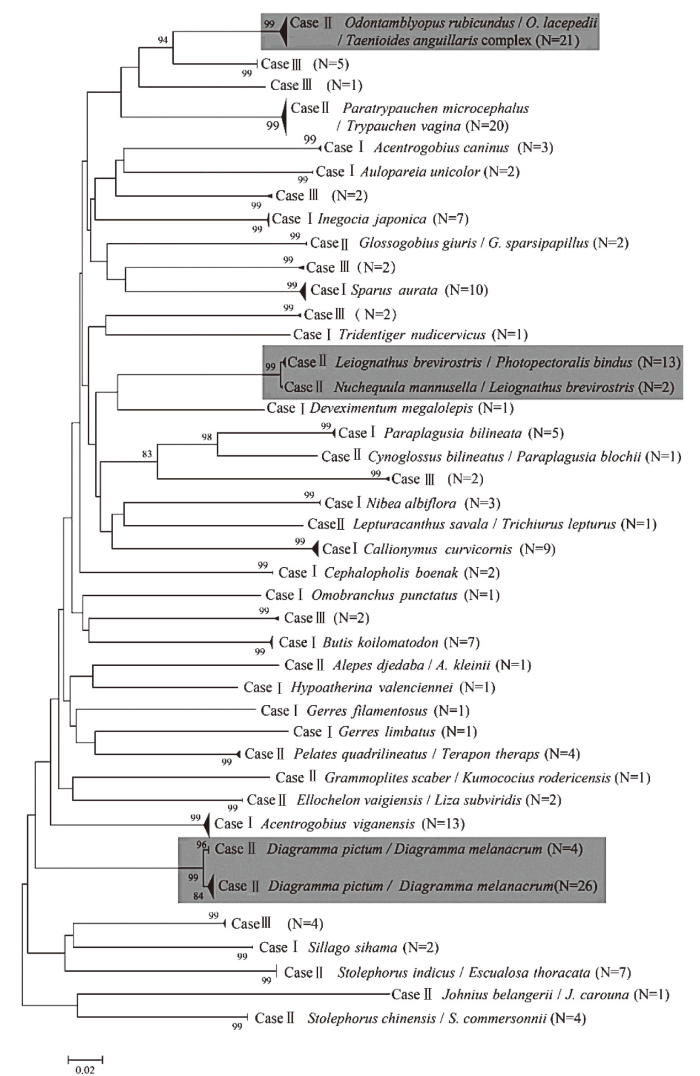
通过BOLD系统序列比对,共界定出43个类群,其中Case Ⅰ(n=69)共界定出17个类群到种水平,占比为39.53%;Case Ⅱ共界定出5个类群到属水平(n=38),11个类群到科水平(n=65)、1个类群到目水平(n=7),占比为39.53%;Case Ⅲ(n=20)共界定出9个类群,未能归类,占比为20.93%(图1、图2)。NJ树分析表明,仔稚鱼类群划分为41个可操作单元(OTUs,图2)。其中,最佳匹配物种红狼牙虾虎鱼(Odontamblyopus rubicundus)和次佳匹配物种雷氏鳗虾虎鱼(O.lacepedii)、鳗虾虎鱼(Taenioides anguillaris)一起形成一个支系(n=21),BOLD数据库中的鱼类种名与序列可能存在分类分歧,该支系仅能界定到虾虎鱼科;少棘胡椒鲷属(Diagramma)(Case Ⅱ)形成独立的2个支系,且节点支持率较高(99%),可能存在潜存种或者错误鉴定。结合BOLD数据库和NJ法,共成功鉴定出32个类群,隶属6目19科20属(表2);未鉴定出9个类群。在32个类群中,2018年夏、秋季鉴定出21个类群(n=95);2019年鉴定出14个类群(n=80);2020年鉴定出3个类群(n=4)。以鲈形目种类最多,共计24个类群,隶属14科16属,数量占比75.88%。科级别以虾虎鱼科种类最多,共计7个类群,数量占比31.16%;其次为鲾科,出现3个类群,数量占比8.04%(表2)。
图2
图2
基于COⅠ基因序列构建的邻接树
Fig.2
The neighbor-joining tree obtained based on COⅠsequences
表2 基于DNA条形码技术鉴定的仔稚鱼类群名录
Tab.2
| 目 Order | 科 Family | 属 Genus | 种/类群 Species /taxa | 样品量 Samples | 年份Year | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2018 | 2019 | 2020 | |||||||||||||||||||
| 鲱形目 Clupeiformes | 鳀科 Engraulidae | 侧带小公鱼属 Stolephorus | 侧带小公鱼属sp. Stolephorus sp. | 4 | 3 | 1 | |||||||||||||||
| 鲱形目sp. Clupeiformes sp. | 7 | 7 | |||||||||||||||||||
| 鲻形目 Mugiliformes | 鲻科 Mugilidae | 鲻科sp. Mugilidae sp. | 2 | 2 | |||||||||||||||||
| 银汉鱼目 Atheriniformes | 银汉鱼科 Atherinidae | 下银汉鱼属 Hypoatherina | 凡氏下银汉鱼 H.valenciennei | 1 | 1 | ||||||||||||||||
| 鲉形目 Scorpaeniformes | 鲬科 Platycephalidae | 瞳鲬属 Inegocia | 日本瞳鲬 I.japonica | 7 | 6 | 1 | |||||||||||||||
| 鲬科sp. Platycephalidae sp. | 1 | 1 | |||||||||||||||||||
| 鲈形目 Perciformes | 鮨科 Serranidae | 九棘鲈属 Cephalopholis | 横纹九棘鲈 C.boenak | 2 | 2 | ||||||||||||||||
| 鱚科 Sillaginidae | 鱚属 Sillago | 多鳞鱚 S.sihama | 2 | 1 | 1 | ||||||||||||||||
| 鲹科 Carangidae | 副叶鲹属 Alepes | 副叶鲹属sp. Alepes sp. | 1 | 1 | |||||||||||||||||
| 鲾科 Leiognathidae | 仰口鲾属 Deveximentum | 大鳞仰口鲾 D.megalolepis | 1 | 1 | |||||||||||||||||
| 鲾科sp.1 Leiognathidae sp.1 | 13 | 13 | |||||||||||||||||||
| 鲾科sp.2 Leiognathidae sp.2 | 2 | 1 | 1 | ||||||||||||||||||
| 银鲈科 Gerreidae | 银鲈属 Gerres | 长棘银鲈 G.filamentosus | 1 | 1 | |||||||||||||||||
| 缘边银鲈 G.limbatus | 1 | 1 | |||||||||||||||||||
| 石鲈科 Pomadasyidae | 少棘胡椒鲷属 Diagramma | 少棘胡椒鲷属sp. Diagramma sp. | 30 | 30 | |||||||||||||||||
| 鲷科 Sparidae | 鲷属 Sparus | 金头鲷 S.aurata | 10 | 8 | 2 | ||||||||||||||||
| 石首鱼科 Sciaenidae | 黄姑鱼属 Nibea | 黄姑鱼 N.albiflora | 3 | 3 | |||||||||||||||||
| 叫姑鱼属 Johnius | 叫姑鱼属sp. Johnius sp. | 1 | 1 | ||||||||||||||||||
| 鯻科 Theraponidae | 鯻科sp. Theraponidae sp. | 4 | 4 | ||||||||||||||||||
| 鳚科 Blenniidae | 肩鳃鳚属 Omobranchus | 斑点肩鳃鳚 O.punctatus | 1 | 1 | |||||||||||||||||
| 䲗科 Callionymidae | 䲗属 Callionymus | 弯棘䲗 C.curvicornis | 9 | 9 | |||||||||||||||||
| 塘鳢科 Eleotridae | 嵴塘鳢属 Butis | 锯嵴塘鳢 B.koilomatodon | 7 | 7 | |||||||||||||||||
| 虾虎鱼科 Gobiidae | 细棘虾虎鱼属 Acentrogobius | 犬牙细棘虾虎鱼 A.caninus | 3 | 2 | 1 | ||||||||||||||||
| 头纹细棘虾虎鱼 A.viganensis | 13 | 13 | |||||||||||||||||||
| 颊沟虾虎鱼属 Aulopareia | 单色颊沟虾虎鱼 A.unicolor | 2 | 2 | ||||||||||||||||||
| 缟虾虎鱼属 Tridentiger | 裸项缟虾虎鱼 T.nudicervicus | 1 | 1 | ||||||||||||||||||
| 舌虾虎鱼属 Glossogobius | 舌虾虎鱼属sp. Glossogobius sp. | 2 | 2 | ||||||||||||||||||
| 虾虎鱼科sp.1 Gobiidae sp.1 | 20 | 20 | |||||||||||||||||||
| 虾虎鱼科sp.2 Gobiidae sp.2 | 21 | 21 | |||||||||||||||||||
| 带鱼科 Trichiuridae | 带鱼科sp. Trichiuridae sp. | 1 | 1 | ||||||||||||||||||
| 鲽形目 Pleuronectiformes | 舌鳎科 Cynoglossidae | 须鳎属 Paraplagusia | 日本须鳎 P.bilineata | 5 | 5 | ||||||||||||||||
| 舌鳎科sp. Cynoglossidae sp. | 1 | 1 | |||||||||||||||||||
| 合计Total | 179 | 95 | 80 | 4 | |||||||||||||||||
3 讨论
3.1 DNA条形码技术在仔稚鱼鉴定中的适用性
条形码思想最早应用于1975年,为美国宾夕法尼亚州大学生物学家丹·詹森对哥斯达黎加西北部的2 000多种昆虫寄生虫进行编目。Amot D E等[21]于1993年首次构思出采用一段DNA序列来标记物种的想法;2002年首次提出了“采用DNA序列作为生物分类系统的主要平台”这一DNA分类学观点[22];2003年,Herbert P D等[23-24]发现利用的一段约648 bp长度的COⅠ基因片段,能够在分子水平上成功地对物种进行区分;2005年,Ward R D等[19]开发的鱼类COⅠ通用引物得到广泛应用,并极大地促进了鱼类相关研究的发展;2006年,MtDNA条形码被首次报道应用于仔稚鱼的鉴定[25]。由于鱼卵、仔稚鱼DNA含量较少,因而采样时容易保存不当,或者提取DNA时出现丢失、断裂和易污染,测序时序列杂峰和简并碱基较多,以及易测序出非目的基因等,鱼卵、仔稚鱼COⅠ基因片段扩增成功率变化较大,在43.2%~80.0% 之间[25⇓⇓⇓-29]。本研究中,COⅠ扩增出的高质量序列占比为84.32%(199/236),低质量序列占比为2.12%(5/236),但是依然有14.57%仔稚鱼没有被成功测序,意味着有更多的仔稚鱼类群有待结合形态学而被鉴定出。基于BOLD比对和NJ法分析,本研究只鉴定出17个类群到种的水平(占比为41.46%),另外有15个物种存在鉴定分歧,仅能鉴定到科、属或者目的水平(Case Ⅱ,占比为36.59%),其可能是存在近期分化的物种或者错误分类所致(图2)。另外,还有9个类群未能通过BOLD系统比对鉴定(占比为21.95%),意味着在研究海域可能还有很多的鱼种缺乏DNA条形码数据库。南海幅员辽阔,地处“珊瑚大三角”西北缘,鱼类生物多样性很高,特有鱼类种类繁多,是世界上鱼类生物多样性热点区域。尽管南海属于西太平洋,但是鱼类区系却属于印度—马来西亚区系,这也导致南海海域与太平洋迥然不同的鱼类区系分布,这在一定程度增加了使用太平洋海域的仔稚鱼书籍鉴定南海的仔稚鱼种类的困难[30⇓⇓-33],而通过分子技术来进行仔稚鱼鉴定,则需要构建一个较为可靠的南海海域的DNA条形码数据库。因而,将DNA条形码技术与传统形态学鉴定相结合,是解决南海鱼卵、仔稚鱼分类鉴定的重要途径。
3.2 徐闻角尾珊瑚礁海域夏、秋季仔稚鱼的种类组成
本研究调查水域主要集中在徐闻珊瑚礁角尾海域,位于琼州海峡最西北侧,紧邻徐闻珊瑚礁国家级保护区。该海域海洋气候条件与紧邻的北部湾相近,属于亚热带—热带海域,鱼类均以暖水性和热水性为主。本研究共成功鉴定出仔稚鱼32个类群,隶属6目19科20属。其中以鲈形目种类最多,共计24个类群,隶属14科16属。科级别以虾虎鱼科种类最多,共计7个类群,到属水平的隶属4属。与杨国欢等[8]先前调查的徐闻珊瑚礁鱼类名录相比,在17个鉴定到种水平的仔稚鱼中,仅有多鳞鱚(Sillago sihama)和长棘银鲈(Gerres filamentosus)2种为共有种,而本研究独自出现的仔稚鱼物种占比达88.23%,意味着徐闻珊瑚礁海域鱼类多样性非常丰富,可能还会有更多的栖息鱼类出现。在本研究中,鉴定的32个类群中,仅横纹九棘鲈(Cephalopholis boenak)等6个种类为礁盘鱼类(Reef-associated species),占比较小(14.63%),未出现隆头鱼科等典型的珊瑚礁鱼类的仔稚鱼。在调查海域,近10年来扇贝类养殖发展迅速,养殖密度很大,海水水质下降,以及活珊瑚覆盖率很低,可能会导致该海域鱼类群落发生改变,珊瑚礁鱼类早期资源补充受到了很大的影响。在未来有待于扩大采样范围,发现更多的仔稚鱼物种,以更全面地反映徐闻角尾海域仔稚鱼的物种多样性。
参考文献
Coral reefs in the South China Sea: their response to and records on past environmental changes
[J].
Confronting the coral reef crisis
[J].
Long-term region-wide declines in Caribbean corals
[J].We report a massive region-wide decline of corals across the entire Caribbean basin, with the average hard coral cover on reefs being reduced by 80%, from about 50% to 10% cover, in three decades. Our meta-analysis shows that patterns of change in coral cover are variable across time periods but largely consistent across subregions, suggesting that local causes have operated with some degree of synchrony on a region-wide scale. Although the rate of coral loss has slowed in the past decade compared to the 1980s, significant declines are persisting. The ability of Caribbean coral reefs to cope with future local and global environmental change may be irretrievably compromised.
Twenty-five years of change in scleractinian coral communities of Daya Bay (northern South China Sea) and its response to the 2008 AD extreme cold climate event
[J].
Long-term decline of a fringing coral reef in the northern South China Sea
[J].
徐闻西岸造石珊瑚礁的组成及空间分布
[J].2008年5–6月在徐闻西岸设置了25个站点, 运用截线样带法调查了造礁石珊瑚的种类多样性和空间分布。本次研究共发现造礁石珊瑚57种, 每个站位的种类数均不超过30种。Shannon-Wiener多样性指数和Pielou均匀度指数均值分别为1.79和0.42。二异角孔珊瑚(Goniopora duofasciata)、盾形陀螺珊瑚(Turbinaria peltata)和锯齿刺星珊瑚(Cyphastrea serailia)是优势种, 其中二异角孔珊瑚为绝对优势种。造礁石珊瑚覆盖率在0–38%之间, 大部分站位介于10–20%, 平均为12.1%。调查区珊瑚礁分布不规则, 呈斑块状分布。
Evaluating the accuracy of morphological identification of larval fishes by applying DNA barcoding
[J].
A checklist of the fishes of the South China Sea
[J].
A checklist of the fishes of southern Taiwan, northern South China Sea
[J].
Identifying the ichthyoplankton of a coral reef using DNA barcodes
[J].Marine fishes exhibit spectacular phenotypic changes during their ontogeny, and the identification of their early stages is challenging due to the paucity of diagnostic morphological characters at the species level. Meanwhile, the importance of early life stages in dispersal and connectivity has recently experienced an increasing interest in conservation programmes for coral reef fishes. This study aims at assessing the effectiveness of DNA barcoding for the automated identification of coral reef fish larvae through large-scale ecosystemic sampling. Fish larvae were mainly collected using bongo nets and light traps around Moorea between September 2008 and August 2010 in 10 sites distributed in open waters. Fish larvae ranged from 2 to 100 mm of total length, with the most abundant individuals being <5 mm. Among the 505 individuals DNA barcoded, 373 larvae (i.e. 75%) were identified to the species level. A total of 106 species were detected, among which 11 corresponded to pelagic and bathypelagic species, while 95 corresponded to species observed at the adult stage on neighbouring reefs. This study highlights the benefits and pitfalls of using standardized molecular systems for species identification and illustrates the new possibilities enabled by DNA barcoding for future work on coral reef fish larval ecology.© 2014 John Wiley & Sons Ltd.
Using DNA barcodes to connect adults and early life stages of marine fishes from the Yucatan Peninsula, Mexico: potential in fisheries management
[J].Barcoding has proven a useful tool in the rapid identification of all life stages of fish species. Such information is of critical importance for fisheries management and conservation, especially in high-diversity regions, such as Mexico’s marine waters, where more than 2200 species occur. The present study reports the barcode analysis of 1392 specimens from the Yucatan Peninsula, corresponding to 610 adults and juveniles, 757 larvae and 25 eggs, representing 181 species (179 teleosts and 2 rays), 136 genera and 74 families. Barcoding results revealed major range extensions and overlooked taxa, including three sympatric species of Albula (one likely undescribed) and a new taxon of Floridichthys. In total, six species of eggs and 34 species of larvae were identified through their barcode match with adults. These cases enabled the first discrimination of the larvae of four species of Eucinostomus, and new information about spawning locality and time was obtained from egg records for the hogfish, Lachnolaimus maximus, which is one of the most commercially important species in the Mexican Caribbean. Also, barcodes revealed mistakes in species recognition during a sport-fish contest. In the future, barcodes will help avoid similar errors and protect rare or endangered species, and will aid regulation of fisheries quotas.
DNA barcoding of freshwater ichthyoplankton in the Neotropics as a tool for ecological monitoring
[J].Quantifying and classifying ichthyoplankton is one of the most effective ways of monitoring the recruitment process in fishes. However, correctly identifying the fish based on morphological characters is extremely difficult, especially in the early stages of development. We examined ichthyoplankton from tributaries and reservoirs along the middle stretch of the Paranapanema River, one of the areas most impacted by hydroelectric projects in the Neotropics. Matching DNA sequences of the COI gene (628-648 bp) allowed us to identify 99.25% of 536 samples of eggs (293) and larvae (243) subjected to BOLD-IDS similarity analysis with a species-level threshold of 1.3%. The results revealed 37 species in 27 genera, 15 families and four orders, some 23.8% of documented fish species in the Paranapanema River. Molecular identification meant that we could include data from egg samples that accounted for about 30% of the species richness observed. The results in this study confirm the efficacy of DNA barcoding in identifying Neotropical ichthyoplankton and show how the data produced provide valuable information for preparing plans for conserving and managing inland waters. © 2015 John Wiley & Sons Ltd.
Quantitative species-level ecology of reef fish larvae via metabarcoding
[J].
Formalin-fixed fish larvae could be effectively identified by DNA Barcodes: a case study on thousands of specimens in South China Sea
[J].Delimiting ichthyoplankton is fundamental work for monitoring the recruitment process and identifying the spawning and nursing grounds of fishes. Nevertheless, it is extremely difficult to identify the fish during the early stages at the species level based on morphological characters because of the paucity of diagnostic features. In this study, we investigated the fish larval community through large-scale ecosystemic sampling in South China Sea (SCS) during 2013 and 2017 using DNA barcodes. To maintain the morphologies of fish larvae, we preserved the larvae in formalin and developed a technique to recover their DNA. Among the 3,500 chosen larvae, we successfully extracted DNA from 2,787 larval samples and obtained 1,006 high-quality sequences. Blast searches showed that 408 larvae (i.e., 40.5%) could be unambiguously identified to species, 413 larvae (i.e., 41.1%) were ambiguously species delimitation, and 185 larvae (i.e., 18.4%) showed a low match similarity with target sequences. A total of 101 species were identified, among which 38 and 33 species corresponded to demersal and reef-associated species, whereas the remaining 30 species corresponded to benthopelagic, pelagic-oceanic, bathypelagic, and pelagic-neritic species. High-quality larval photographs of the 101 diagnosed species showed intact morphological characters and thus provided a reference for identifying fish species during the early stages based on morphological characters. Our study highlighted the possibility of recovering and amplifying DNA from formalin-fixed samples and provided new insight into the fish larval community in the SCS.
Bold: the barcode of life data system
[J].
DNA barcoding Australia’s fish species
[J].
A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences
[J].Some simple formulae were obtained which enable us to estimate evolutionary distances in terms of the number of nucleotide substitutions (and, also, the evolutionary rates when the divergence times are known). In comparing a pair of nucleotide sequences, we distinguish two types of differences; if homologous sites are occupied by different nucleotide bases but both are purines or both pyrimidines, the difference is called type I (or "transition" type), while, if one of the two is a purine and the other is a pyrimidine, the difference is called type II (or "transversion" type). Letting P and Q be respectively the fractions of nucleotide sites showing type I and type II differences between two sequences compared, then the evolutionary distance per site is K = -(1/2) ln [(1-2P-Q) square root of 1-2Q]. The evolutionary rate per year is then given by k = K/(2T), where T is the time since the divergence of the two sequences. If only the third codon positions are compared, the synonymous component of the evolutionary base substitutions per site is estimated by K'S = -(1/2) ln (1-2P-Q). Also, formulae for standard errors were obtained. Some examples were worked out using reported globin sequences to show that synonymous substitutions occur at much higher rates than amino acid-altering substitutions in evolution.
Digital codes from hypervariable tandemly repeated DNA sequences in the Plasmodium falciparum circumsporozoite gene can genetically barcode isolates
[J].
DNA points the way ahead in taxonomy
[J].
Biological identifications through DNA barcodes
[J].
Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species
[J].
MtDNA barcode identification of fish larvae in the southern Great Barrier Reef, Australia
[J].
DNA条形码在厦门湾鱼卵和仔稚鱼分类鉴定中的应用
[J].本研究选取2017年4月采集的厦门湾鱼卵、仔稚鱼样品进行DNA 条形码分析。获得鱼卵、仔稚鱼的COI 基因序列27 条, 共鉴定种类11种,全部鉴定到种,隶属于4目9科11 属。在系统进化树的分析中, 同一种类的不同个体都聚在一支, 所有物种都分别聚为独立的一支, 这些物种都能有效地区分开来,说明利用COI基因可以进行有效的仔稚鱼物种鉴定。同时获取11种鱼类浮游生物高清图像,并描述其甄别性的形态特征。以上结果表明, 线粒体COI基因作为DNA 条形码可以实现鱼卵和仔稚鱼的物种鉴定, 且较多能鉴定到种的水平。
基于CO Ⅰ和16S rRNA基因片段鉴定厦门海域的仔稚鱼
[J].本文基于线粒体细胞色素C氧化酶亚基 I(COI)基因和16S rRNA基因片段对采集于厦门海域的仔稚鱼样品进行种类鉴定,探究其在仔稚鱼种类鉴定中的适用性。研究共获得64条COI基因序列和74条16S rRNA基因序列,通过序列比对,COI基因将仔稚鱼样品鉴定为26个种类,其中19个种类鉴定到种、6个鉴定到属、1个种类仅鉴定到科;16S rRNA基因将仔稚鱼样品鉴定为29个种类,其中23个种类鉴定到种、6个鉴定到属。COI基因的平均种内遗传距离为0.001 5,平均种间遗传距离为0.197 6,16S rRNA基因的平均种内遗传距离为0.000 3,平均种间遗传距离为0.089 2,COI和16S rRNA基因的平均种间遗传距离都为平均种内遗传距离的10倍以上,两者都可以进行有效的仔稚鱼种类鉴定。在基于COI和16S rRNA基因构建的系统进化树上,所有物种都分别单独聚为一支,同一个种类的不同个体都能聚在同一个分支,这些物种均能得到有效区分。以上结果表明,COI和16S rRNA基因均可以实现仔稚鱼的种类鉴定,两种基因结合使用可以提高仔稚鱼种类鉴定的准确性。
Assemblage structure of the ichthyoplankton and its relationship with environmental factors in spring and autumn off the Pearl River Estuary
[J].Ichthyoplankton assemblages and their relationship with environmental variables are investigated in waters off the Pearl River Estuary in spring and autumn of 2019. Of 80 ichthyoplankton taxa identified using DNA barcode and morphological methods, 61 are identified to species. The most abundance families (Carangidae, Trichiuridae, Mullidae, and Scombridae) account for 61.34% of the horizontal total catch in spring, while Menidae and Carangidae are the most abundant families identified in autumn, accounting for 89.72% of the horizontal total catch. Cluster analysis identifies three species assemblages in spring, and four in autumn based on horizontal trawls. Relationships between assemblage structure and environmental variables (in situ and remote sensed) are determined by canonical correspondence analysis. Ichthyoplankton assemblage structure appears to be strongly influenced by sea level anomalies, salinity, water depth, temperature at 10 m depth, and distance from shore. We demonstrate the efficacy of using DNA barcode to identify ichthyoplankton, and suggest how these data can be used to protect fish spawning grounds in waters off the Pearl River Estuary.